VMTK-Neuro: Visual Manipulation Toolkit on NeuroImages
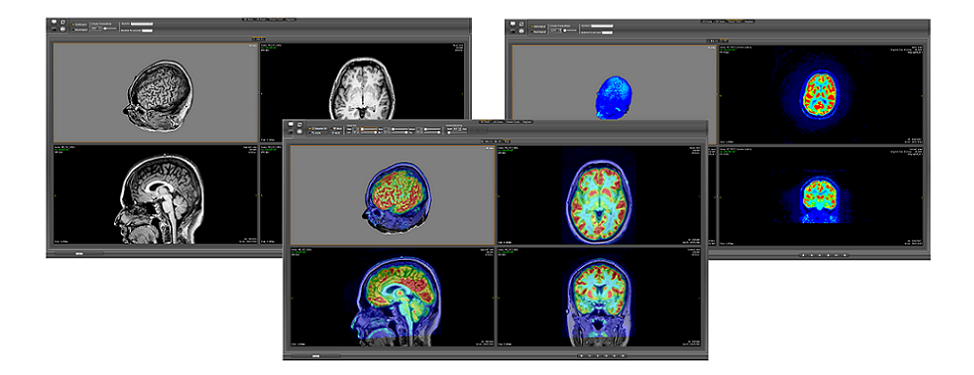
VMTK-Neuro was born from the idea of developing an application that allows neurologists to non-invasively explore a brain to identify subtle structural abnormalities, such as dysplastic lesions. Features, such as patient-oriented slice views, views of fused multimodality data, overview-plus-detail visualization, multiplanar reformatting, and curvilinear reformatting, are integrated to provide a coordinated view of the contents of 2D and 3D multiple views of a brain. The keys to our solution are:
1. To distinguish five reference spaces along the data processing from the raw data (in DICOM format) to the format of the displayed images. They are Patient-oriented coordinate reference (DCR), native coordinate reference (PhCR), normalized native coordinate reference (NPhCR), and texture coordinate reference (TCR).
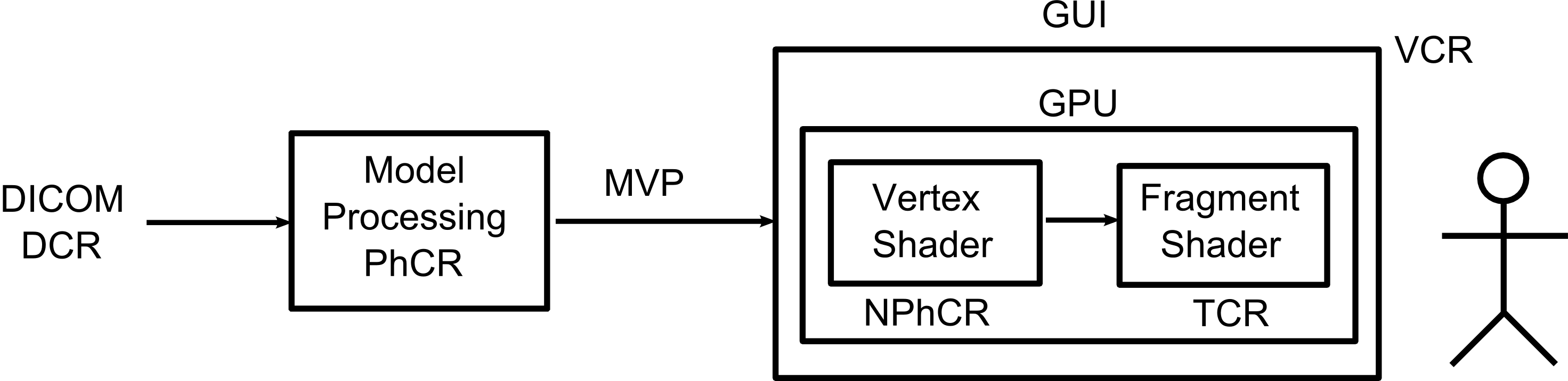
2. To move most of the interactive rendering algorithms to GPU to reduce CPU-to-GPU data transfer latency. The raw data are loaded in the GPU memory with texture coordinates (TCR) and different operations on the data volume are carried directly in the TCR space. Only the updated control data Ω and the viewing data MVP are continually resent to GPU during user interactions.
3. To generate ahead the data that are necessary for accomplishing smooth interactions. Any visible voxel of the displayed image is an active blobby that can readily react in the PhCR space while interacting with the displayed image.
4. To integrate human reasoning in a volume processing pipeline that contains stages classified as open problems. User-friendly interfaces are provided to contextualize the processing phase to an expert and to guide her/im helping the computer accomplish a task.
It has evolved into an application that supports neurosurgical planning of the resection of cortical lesions in version 3.2. The cortical lesions are structural abnormalities seated in the outermost sheet of the brain. The curvilinear reformatting of the T1-weighted magnetic resonance images with superimposed superficial veins provides unique contextualized views of the lesions to be resected. The superficial veins are visible in gadolinium contrast-enhanced T1-weighted magnetic resonance images.
It has evolved into an application that supports neurosurgical planning of the resection of cortical lesions in version 3.2. The cortical lesions are structural abnormalities seated in the outermost sheet of the brain. The curvilinear reformatting of the T1-weighted magnetic resonance images with superimposed superficial veins provides unique contextualized views of the lesions to be resected. The superficial veins are visible in gadolinium contrast-enhanced T1-weighted magnetic resonance images.
The functions developed so far opened up different perspectives for applications. However, restructuring the architecture was necessary to allow their sharing. On top of the restructured VMTK-Neuro, we may develop independently deployable applications. This version developed three application prototypes in parallel: Neuroimage-guided Placement of Sensors on a Head, Automated Anatomical Labeling of Gross Neuroanatomy, and Visual Analytics of Neural Connections for Neurosurgical Planning.
Common features:
1) Multi-windows
2) Co-registration
3) Color map editor
4) Pointing device (mouse)
5) Multiplanar Reformatting
6) Curvilinear Reformatting
7) Multimodal Visualization
8) 2D and 3D synchronized slicing
Application Prototypes:
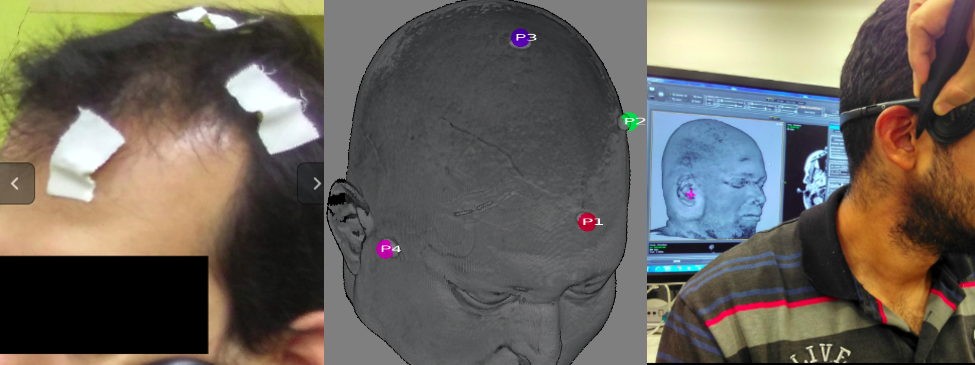
Neuroimage-guided Placement of Sensors on a Head: For supporting a more accurate placement of sensors (electrodes and optodes) on a head, this application provides an interactive environment for co-registering a real human head to the corresponding magnetic resonance imaging of the head. The positions digitized by the Polhemus Fastrack Digitizer are input through the serial port RS-232 and converted to the patient-oriented coordinates to be visually fed back on the displayed head's image.
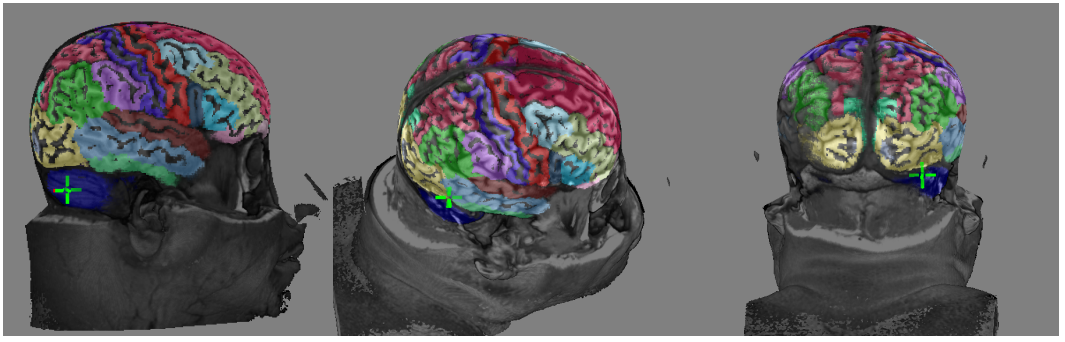
Automated Anatomical Labeling of Gross Neuroanatomy: For supporting the teaching of gross neuroanamoty, this application provides an interactive environment for labeling a user-specified brain based on the Talairach digital atlas (Talairach space) and MNI-ICB2009c atlas (MNI space).
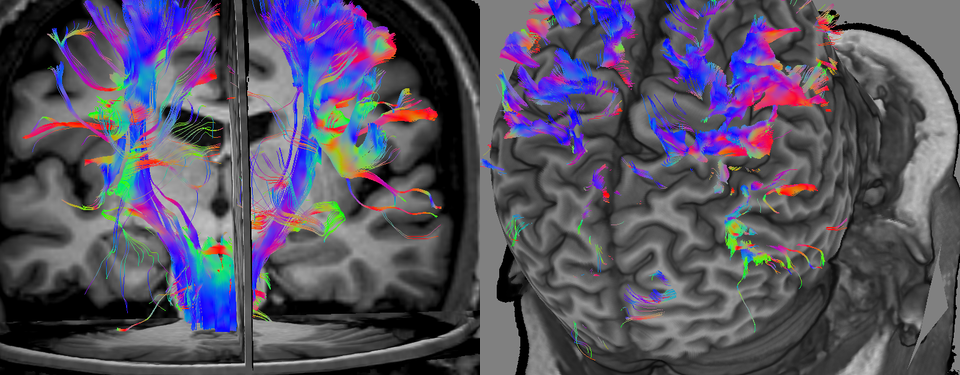
Visual Analytics of Neural Connections for Neurosurgical Planning: For supporting preoperative spatial analysis of the vicinity of a resection cavity of a lesion relative to the eloquent fiber tracts, this application provides several rendering modes of diffusion data over a T1-weighted magnetic resonance image. They range from diffusivity maps and diffusion direction glyphs to tractography. A user can inspect these data from various views at an interactive rate.