Mestrado Dissertation's Adviser: Profa. Dr-Ing.Wu, Shin-Ting
Although the medical scanning technology (e.g. CT, MRI, PET and SPECT) has evolved rapidly over the past years, new visualization algorithms were devised and the existing ones were improved, the interactive visualization techniques which enable physicians to gain insight into complex internal human structure is still at its very beginning stage and is a challenging research field.
The Focus+context visualization and volume clipping techniques are representative efforts towards an interactive environment that facilitate medical diagnosis. Multimodal rendering, multi-dimensional transfer functions and magic volume lenses have been proposed to emphasize important regions of volumetric data (focus), and deemphasize their surroundings (context). Also, several clipping algorithms have been developed to selectively remove parts of volume data for revealing and exploring hidden regions (e.g. axis-aligned and oblique cuts, cuts in pre-specified arbitrary geometry and curvilinear cut-sections parallel to the head’s scalp). But, concerning dynamic queries that continuously update the filtered data while the user hovers the cursor over the region of interest, the results are not entirely satisfactory. The basic premise in query-driven visualization is its capability in limiting visualization and analytics processing the data that are “important” to the users.
The proposal for this master thesis is the development of dynamic query tools for exploring 3D medical images interactively. Our main goal is to help neurosurgeons and physicians detect cortical lesions and plan surgery of removal or disconnection of the abnormal cortical tissue from the rest of brain with minimal damage.
We present three dynamics query tools: the 3D cursor, the volumetric probe and the moving magnifying lens. The first one provides a very intuitive way to explore each volume sample. The second tool allows selecting and highlighting features of interest with pre-defined one-dimensional transfer function, while the third one magnifies regions over which the cursor pointer is hovered. The main difference of our proposal is that the user may manipulate the displayed objects as if it were in her/is hands. The cursor pointer can be arbitrarily moved in 3D, either on the screen plane or entering/leaving it.
To achieve interactive time performance, we also propose to explore the GPU SIMD-processing capability by using non-displayable OpenGL framebuffer object to transfer non-graphics data from/to GPU.

We applied our methods to MRI images provided by our university hospital.
A short video as illustration about how to use the 3D cursor is presented below.
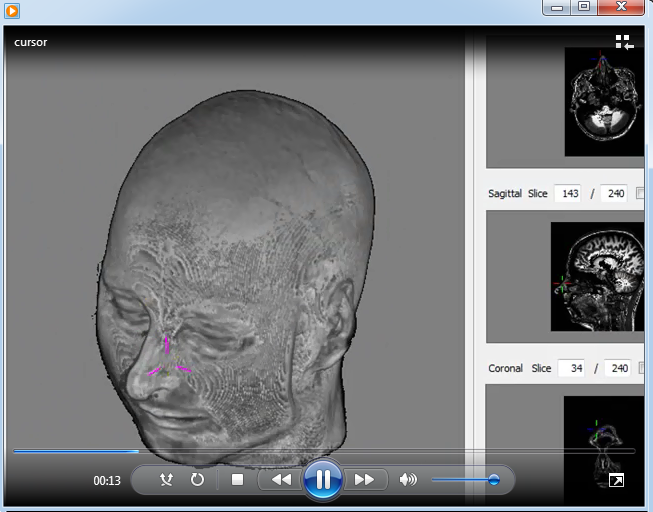 |
This video shows how the use the 3D Cursor. MP4 (2.9MB) |
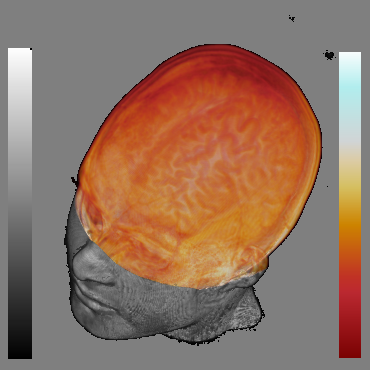
Fig. 2 shows thefocal spherical region located over the brain. It allows distinguishthe brain surface. As the rendering of both focal and context regionsis performed in the same pass on current GPUs, our system is veryfast.
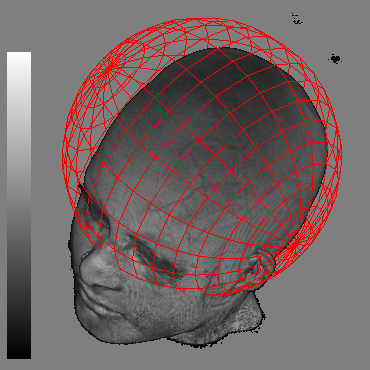 | 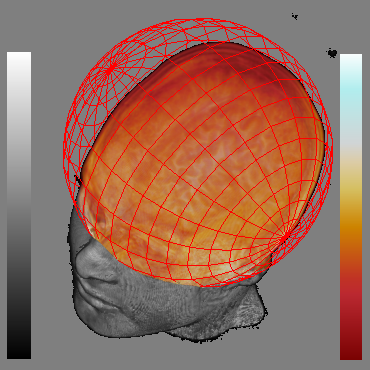 |  |
| (a) | (b) | (c) |
An instructional video about how to use the probe tool is presented below:
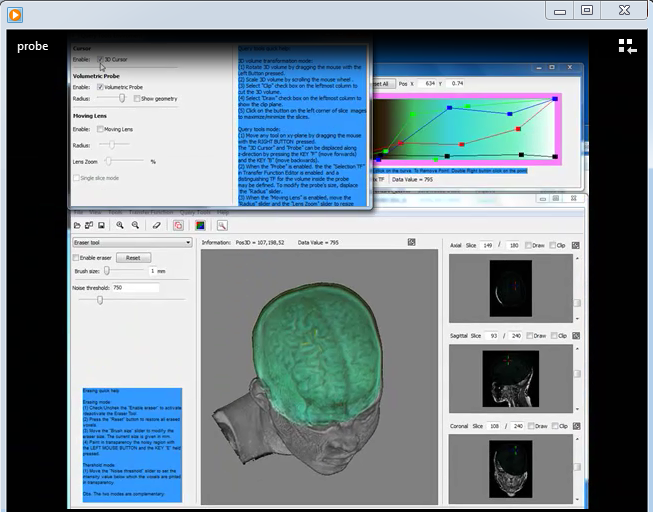 |
This video shows how to use the volumetric probe. MP4 (7. 76MB) |
Our lens is distinguished from otherapproaches in that it is configurable to only show the colors attributed to the closest visible voxels (non-transparent voxel). The alpha blending of all colors of the voxels along each viewing ray traversal is an alternative mode, in consequence, the outcome may be less blurred.
Fig. 3 illustrates the outcome of our proposed tool with respect to other alternatives.
 | 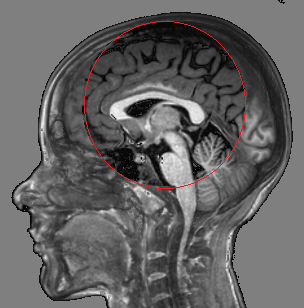 | 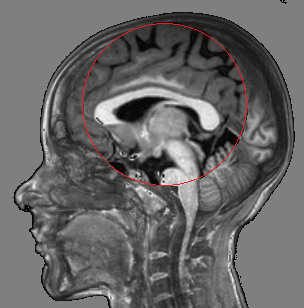 | 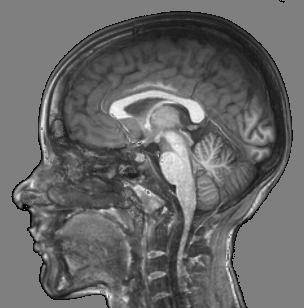 |
| (a) | (b) | (c) | (d) |
An instructional video about our moving lens is presented below:
 |
This video shows how to use the moving magnification lens. MP4 (4. 76MB) |
This project was supported by CAPES (Coordination for the Improvement of Higher Level Personel). Without medical constructive feedback of Dr. Clarissa Lin Yasuda, Dr. Ana Carolina Coan and Prof. Dr. Fernando Cendes along the development of this project, we would not have achieved the results presented.
The prototype was implemented in C++ with a volume ray-casting fragment shader written in GLSL. The wxWidgets GUI library was used for implementing the interface and the open source Grassroots DiCoM library for reading and parsing Dicom medical files. The documentation of the source code of this tool was generated by doxygen. Its complete online version is accessible from here.
Here you can download a trial version of our software.
My master thesis in portuguese is available in pdf.
Requirement The basic requirement of graphics hardware is that supports both OpenGL and GLSL 3.0 or higher. We recommend a video memory of 1.0 GB.